Integrating multiple references for single-cell assignment
Abstract
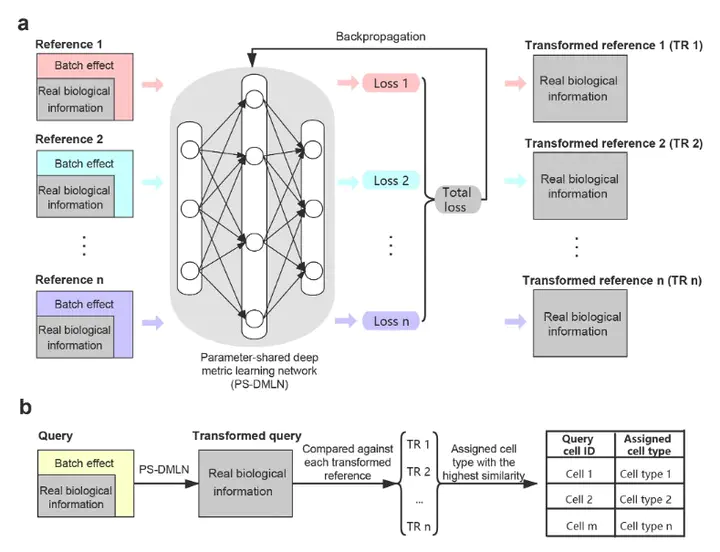
Efficient single-cell assignment is essential for single-cell sequencing data analysis. With the explosive growth of single-cell sequencing data, multiple single-cell sequencing data sources are available for the same kind of tissue, which can be integrated to further improve single-cell assignment; however, an efficient integration strategy is still lacking due to the great challenges of data heterogeneity existing in multiple references. To this end, we present mtSC, a flexible single-cell assignment framework that integrates multiple references based on multitask deep metric learning designed specifically for cell type identification within tissues with multiple single-cell sequencing data as references. mtSC github.
Supplementary notes can be added here, including code and math.