Learning for single-cell assignment
Abstract
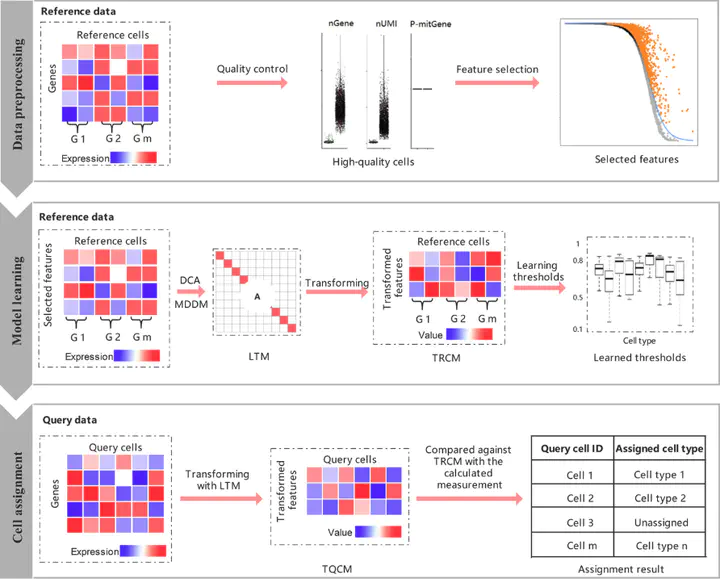
Efficient single-cell assignment without prior marker gene annotations is essential for single-cell sequencing data analysis. Current methods, however, have limited effectiveness for distinct single-cell assignment. They failed to achieve a well-generalized performance in different tasks because of the inherent heterogeneity of different single-cell sequencing datasets and different single-cell types. Furthermore, current methods are inefficient to identify novel cell types that are absent in the reference datasets. To this end, we present scLearn, a learning-based framework that automatically infers quantitative measurement/similarity and threshold that can be used for different single-cell assignment tasks, achieving a well-generalized assignment performance on different single-cell types. We evaluated scLearn on a comprehensive set of publicly available benchmark datasets. scLearn github.
Supplementary notes can be added here, including code and math.